Predicting bacterial transcription factor binding sites through machine learning and structural characterization based on DNA duplex stability
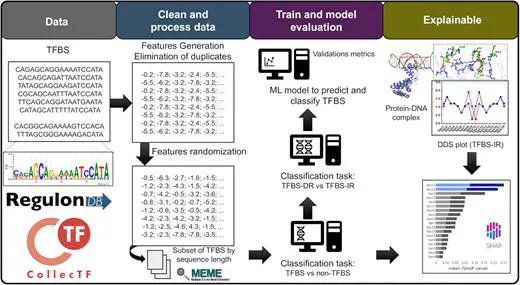 Schematic illustrating the workflow for acquiring, training, validating, and interpreting the predictive model designed to TFBS
Schematic illustrating the workflow for acquiring, training, validating, and interpreting the predictive model designed to TFBSResumen
Transcriptional factors (TFs) in bacteria play a crucial role in gene regulation by binding to specific DNA sequences, thereby assisting in the activation or repression of genes. Despite their central role, deciphering shape recognition of bacterial TFs-DNA interactions remains an intricate challenge. A deeper understanding of DNA secondary structures could greatly enhance our knowledge of how TFs recognize and interact with DNA, thereby elucidating their biological function. In this study, we employed machine learning algorithms to predict transcription factor binding sites (TFBS) and classify them as directed-repeat (DR) or inverted-repeat (IR). To accomplish this, we divided the set of TFBS nucleotide sequences by size, ranging from 8 to 20 base pairs, and converted them into thermodynamic data known as DNA duplex stability (DDS). Our results demonstrate that the Random Forest algorithm accurately predicts TFBS with an average accuracy of over 82% and effectively distinguishes between IR and DR with an accuracy of 89%. Interestingly, upon converting the base pairs of several TFBS-IR into DDS values, we observed a symmetric profile typical of the palindromic structure associated with these architectures. This study presents a novel TFBS prediction model based on a DDS characteristic that may indicate how respective proteins interact with base pairs, thus providing insights into molecular mechanisms underlying bacterial TFs-DNA interaction.